All MCAT Biology Resources
Example Questions
Example Question #22 : Dna, Rna, And Proteins
Prokaryotic mRNA usually includes several genes on the same transcript. An operon is a genetic unit which typically consists of a promoter, an operator, and all of the functional genes that will be coded for by a single mRNA. In biology, the lac operon is the most commonly used example. The lac operon is transcribed when a prokaryote has a glucose deficiency, and requires lactose in order to create glucose.
The lac operon is modulated by specific proteins which can regulate the amount of transcription. A lac repressor will attach to the operator and reduce transcription if it is not necessary at a given time. The lac repressor will be removed in the presence of lactose by becoming attached to a lactose molecule. Meanwhile, catabolite activator protein (CAP) will attach upstream to the promoter and signal RNA polymerase to begin transcription if transcription is needed.
Imagine that there is a mutation on the lac repressor which makes it unable for lactose to attach. What would you expect to happen?
CAP will be unable to attach to the promoter
The lac repressor will be unable to attach to the operator
Lac operon transcription will be very low
Lac operon transcription will no longer be inhibited
Lac operon transcription will be very low
The lac repressor is attached to the operator in the absence of lactose; it will only be removed from the operator if lactose attaches to it. Since there is a mutation that forbids this attachment, the lac repressor will always be on the operator. This will result in very low lac operon transcription, even in the presence of lactose.
Catabolite activator protein (CAP) will still bind upstream of the promoter, but RNA polymerase will be unable to access the operon due to the repressor's presence.
Example Question #11 : Transcription
Although there are about 30,000 genes in the human body, approximately 120,000 proteins can be created in the body. This is caused by __________.
the polycistronic transcription mechanism found in humans
the degeneracy fo the genetic code
the variable splicing of exons from pre-mRNA
the addition of a poly A tail to the primary transcript
the variable splicing of exons from pre-mRNA
The primary transcript (immediately following transcription) is composed of multiple segments of RNA called introns and exons. While introns are always cleaved from the primary transcript, the exons may or may not stay on the primary transcript, which will eventually be translated. Since different numbers and orders of exons can arise from the same transcript, there are multiple proteins that can possibly be made from one primary transcript.
Intron and exon splicing, the addition of the 5' cap, and the addition of the poly A tail are all crucial post-transcriptional modifications in eukaryotes, but only the alternative splicing patterns result in variable transcripts. Eukaryotes do not follow the polycistronic model, in which one mRNA will code for multiple genes or proteins. The degeneracy of the genetic code refers to the multiple codons that will produce the same amino acids; this feature increases the variability of the DNA sequence, but does not affect the number of possible transcripts, genes, or proteins formed.
Example Question #131 : Cell Biology, Molecular Biology, And Genetics
What is the purpose of the poly A tail attached to the primary transcript prior to translation?
It signals the ribosome to terminate translation
It protects the transcript from degradation
It gives the ribosome a site to begin translation
It signals the mRNA where to travel in order to be translated
It protects the transcript from degradation
The poly A tail is an example of post-transcriptional processing. The poly A tail is attached to the end of the transcript, and protects it from being degraded by exonucleases. Exonucleases are essential in order to recycle the nucleobases used in mRNA, but can be harmful to RNA that has not yet been translated unless the poly A tail is present.
Example Question #11 : Transcription
What is a common modification that occurs on histone tails to modulate gene expression?
Sumoylation
Ubiquitination
Hydroxylation
Acetylation
Acetylation
Both acetylation and methylation of histone tails modulate expression of target genes. Acetylation is always associated with active transcription while, in most cases, methylation is associated with repression of target genes. Acetylation generally loosens the DNA-histone association, allowing for transcription. In contrast, methylation generally tightens this association, blocking transcription proteins from binding to the DNA.
Hydroxylation is used to modify proteins and activate steroid hormones. Ubiquitination and sumoylation are post-translation protein modifications.
Example Question #11 : Transcription
Which of the following experimental methods cannot be used to measure the relative abundance of a particular mRNA?
Northern blot
Reverse transcription polymerase chain reaction (rt-PCR)
Microarray
Western blot
Western blot
Western blots are used to measure the relative abundance of proteins. While there is a correlation between the amount of mRNA and protein, using a western blot to measure mRNA would be inconclusive due to the variability of protein half life.
Northern blots are used to run mRNA samples on gels, DNA microarrys give the expression levels of certain genes, and rtPCR is used to detect RNA expression levels. Any of these methods could provide the relative abundance of a particular mRNA.
Example Question #11 : Transcription
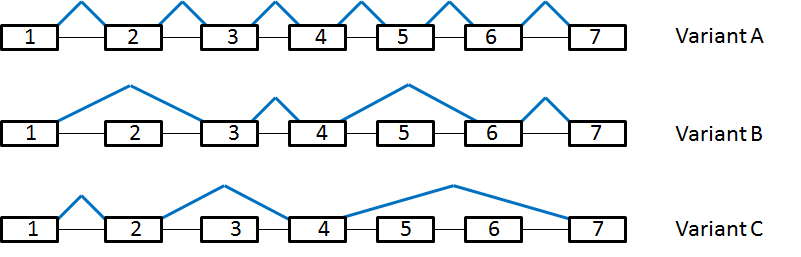
Three isoforms of a particular protein (variants A, B, and C) are expressed. In the following diagram, exons are indicating by boxes, introns by the black lines, and splice junctions by blue lines. Assume each exon is greater than 500 bp in length (figure not drawn to scale).
In the absence of available antibodies for Western blotting, how would one experimentally determine if variant C was expressed in HeLa cells?
Quantitative real time PCR using primers to amplify exon 6
Quantitative real time PCR using primers to amplify exon 2
Quantitative real time PCR using primers to amplify exon 1
Quantitative real time PCR using primers designed to amplify a region from the 3' end of exon 1 to the 5' end of exon 2
Quantitative real time PCR using primers designed to amplify a region from the 3' end of exon 4 to the 5' end of exon 7
Quantitative real time PCR using primers designed to amplify a region from the 3' end of exon 4 to the 5' end of exon 7
Although each splice variant contains exons 4 and 7, PCR products between these exons in variants A and B would be too large to be detected using real time PCR; therefore, amplifying that region would conclusively identify the presence of variant C in HeLa cells. If variant C is not expressed in HeLa cells, one would expect a negative (or unamplified) result using real time PCR.
Variant A expression: 1, 2, 3, 4, 5, 6, 7
Variant B expression: 1, 3, 4, 6, 7
Variant C expression: 1, 2, 4, 7
By amplifying exons 5 and 6, and looking for a negative result, we can identify the presence of variant C.
Certified Tutor
All MCAT Biology Resources