All MCAT Biology Resources
Example Questions
Example Question #5 : Structure Of Dna And Rna
Scientists use a process called Flourescent In-Situ Hybridization, or FISH, to study genetic disorders in humans. FISH is a technique that uses spectrographic analysis to determine the presence or absence, as well as the relative abundance, of genetic material in human cells.
To use FISH, scientists apply fluorescently-labeled bits of DNA of a known color, called probes, to samples of test DNA. These probes anneal to the sample DNA, and scientists can read the colors that result using laboratory equipment. One common use of FISH is to determine the presence of extra DNA in conditions of aneuploidy, a state in which a human cell has an abnormal number of chromosomes. Chromosomes are collections of DNA, the totality of which makes up a cell’s genome. Another typical use is in the study of cancer cells, where scientists use FISH labels to ascertain if genes have moved inappropriately in a cell’s genome.
Using red fluorescent tags, scientists label probe DNA for a gene known to be expressed more heavily in cancer cells than normal cells. They then label a probe for an immediately adjacent DNA sequence with a green fluorescent tag. Both probes are then added to three dishes, shown below. In dish 1 human bladder cells are incubated with the probes, in dish 2 human epithelial cells are incubated, and in dish 3 known non-cancerous cells are used. The relative luminescence observed in regions of interest in all dishes is shown below.
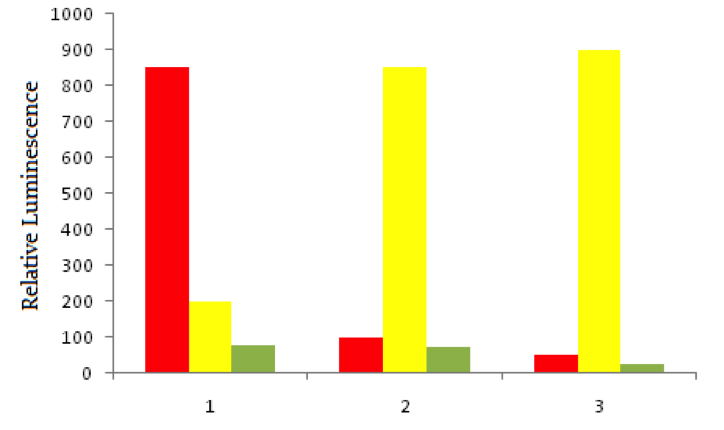
When probe DNA binds with target DNA, what is the main bonding mechanism likely at play?
Hydrogen bonding
Ionic bonding
Coordinate covalent bonding
Covalent bonding
Van der Waal forces
Hydrogen bonding
When two complimentary strands of nucleic acid bind together, hydrogen bonding is the bonding at play. Guanine-cytosine pairs have three hydrogen bonds between them, and adenine-thymine pairs have two.
Example Question #1 : Nucleic Acids
In which of the following ways does RNA differ from DNA?
The sugar components are different
RNA has the base uracil instead of adenine
Proteins do not form from RNA
The phosphate groups are different
The sugar components are different
RNA contains a ribose sugar, while DNA contains a deoxyribose sugar (missing a hydroxyl group at the second carbon). The phosphate groups are the same in both molecules, and RNA replaces the base thymine, not adenine, with uracil.
Example Question #7 : Structure Of Dna And Rna
Which of the following is NOT found in both RNA and DNA?
Nitrogenous bases
A pentose sugar
Phosphodiester bonds
Thymine
Thymine
RNA and DNA share many common attributes. The nucleotides are attached to one another via phosphodiester bonds. They both have a pentose sugar, as well as nitrogenous bases. Only DNA, however, has thymine bases. RNA uses uracil in place of thymine.
Example Question #2 : Nucleic Acids
Which of the following statements is not true about DNA and RNA?
Both DNA and RNA have thymine bases
Both DNA and RNA are polymerized with phosphodiester bonds
Both DNA and RNA have adenine bases
Both DNA and RNA have a pentose sugar
Both DNA and RNA have thymine bases
DNA and RNA differ in the nitrogenous bases used in their nucleotides; DNA uses thymine, while RNA uses uracil. Both nucleic acids are joined into polymers by phosphodiester bonds and use adenine as a nitrogenous base. Both contain a pentose sugar: DNA uses deoxyribose, while RNA uses ribose.
Example Question #1 : Structure Of Dna And Rna
DNA is comprised of a double-stranded helix in which purine bases are paired with pyrimidine bases. Which base pairing requires more energy to separate?
Adenine and thymine because they are paired by three hydrogen bonds
Adenine and thymine because they are paired by covalent bonds
Guanine and cytosine because they are paired by three hydrogen bonds
Guanine and cytosine because they are paired by covalent bonds
Guanine and cytosine because they are paired by three hydrogen bonds
All purine-pyrimindine pairs are bound by hydrogen bonds; covalent bonds are only found in the DNA backbone. Guanine-cytosine pairs are bound by thee hydrogen bonds, while adenine-thymine pairs are bound by two hydrogen bonds. The additional bond adds additional stability and energy to the guanine-cytosine linkage, making it harder to separate this base pair.
Example Question #1 : Nucleic Acids
Hypersensitivity reactions occur when body tissues are affected by an abnormal immune reaction. The result is damage to normal tissues and clinical illness. A peanut allergy is an example of a hypersensitivity reaction, but there are three additional broad classes.
One class involves the abnormal production or deposition of antibodies. Antibodies are B-cell derived molecules that normally adhere to pathogens, rendering them unable to continue an infection. When antibodies are produced against normal tissues, however, disease can result. Figure 1 depicts a schematic structure of an antibody.
Antibodies can be divided into two peptide chains: heavy and light. Heavy chains form the backbone of the antibody, and are attached to light chains via covalent bonding. Each heavy and light chain is then further divided into constant and variable regions. Variable regions exhibit molecular variety, generating a unique chemical identity for each antibody. These unique patterns help guarantee that the body can produce antibodies to recognize many possible molecular patterns on invading pathogens.
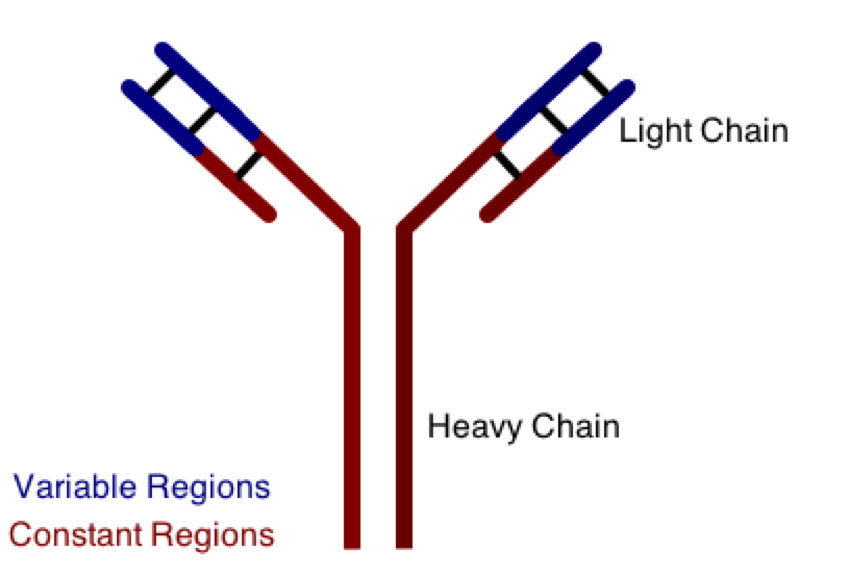
Humans must generate an enormous array of antibodies to account for all the possible patterns that they must recognize on pathogens. In fact, the diversity needed in antibody chains cannot be explained by DNA sequence variation. As a result, segments of DNA are physically rearranged in a process called VDJ rearrangement in order to create unique antibody chains. In order to carry out this process, DNA bonds must be broken. What kinds of intra-strand DNA bonds are most likely broken in this process?
Hydrogen bonds
van der Waals interactions
Phosphodiester covalent bonds
Peptide bonds
Base stacking interactions
Phosphodiester covalent bonds
Intra-strand DNA bonds are bonds that exist within each of the two complementary strands that make up a DNA molecule. The two strands are held together by hydrogen bonds between nitrogenous bases and base stacking interactions between the aromatic rings of the bases. Phosphodiester bonds tie together the adjacent ribose sugars in the DNA backbone, and thus give structure to each DNA strand.
Essentially, phosphodiester bonds are the primary intra-strand linkage, while hydrogen bonds are the primary inter-strand linkage.
Example Question #11 : Structure Of Dna And Rna
In 2013, scientists linked a cellular response called the unfolded protein response (UPR) to a series of neurodegenerative diseases, including such major health issues as Parkinson’s and Alzheimer’s Disease. According to their work, the unfolded protein response is a reduction in translation as a result of a series of enzymes that modify a translation initiation factor, eIF2, as below:
![]()
In the above sequence, the unfolded protein sensor binds to unfolded protein, such as the pathogenic amyloid-beta found in the brains of Alzheimer’s Disease patients. This sensor then phosphorylates PERK, or protein kinase RNA-like endoplasmic reticulum kinase. This leads to downstream effects on eIF2, inhibition of which represses translation. It is thought that symptoms of neurodegenerative disease may be a result of this reduced translation.
During translation, the genetic code is used to convert a sequence of nitrogenous bases in mRNA to an amino acid sequence. Which of the following is a difference between the mRNA transcript and the original DNA?
I. DNA has no hydroxol group at the sugar's 2' position
II. DNA has thymine; RNA has uracil
III. DNA is synthesized 5' to 3'; RNA is synthesized 3' to 5'
III, only
I and II
II and III
I, only
I, II, and III
I and II
DNA and RNA differ mainly due to the missing hydroxol group, and the replacement of thymine with uracil in the latter. Both are synthesized 5' to 3'.
Example Question #1 : Electrochemical, Environmental, And Other Analyses
Once a sample of DNA is isolated, it is loaded on an agarose electrophoresis gel, as shown below. Once the sample has run, where will the student find the DNA?
It is impossible to tell without knowing the charge of the DNA
Red section
It is impossible to tell without knowing the size of the DNA fragments
Green section
Green section
DNA is negatively charged, so it will migrate toward the positive electrode during electrophoresis. As a result, it will migrate from the center line into the green region of the gel.
Example Question #11 : Structure Of Dna And Rna
Prions are the suspected cause of a wide variety of neurodegenerative diseases in mammals. According to prevailing theory, prions are infectious particles made only of protein and found in high concentrations in the brains of infected animals. All mammals produce normal prion protein, PrPC, a transmembrane protein whose function remains unclear.
Infectious prions, PrPRes, induce conformational changes in the existing PrPC proteins according to the following reaction:
PrPC + PrPRes → PrPRes + PrPRes
The PrPRes is then suspected to accumulate in the nervous tissue of infected patients and cause disease. This model of transmission generates replicated proteins, but does so bypassing the standard model of the central dogma of molecular biology. Transcription and translation apparently do not play a role in this replication process.
This theory is a major departure from previously established biological dogma. A scientist decides to test the protein-only theory of prion propagation. He establishes his experiment as follows:
Homogenized brain matter of infected rabbits is injected into the brains of healthy rabbits, as per the following table:
Rabbit 1 and 2: injected with normal saline on days 1 and 2
The above trials serve as controls.
Rabbit 3 and 4: injected with homogenized brain matter on days 1 and 2
The above trials use unmodified brain matter.
Rabbit 5 and 6: injected with irradiated homogenized brain matter on days 1 and 2
The above trials use brain matter that has been irradiated to destroy nucleic acids in the homogenate.
Rabbit 7 and 8: injected with protein-free centrifuged homogenized brain matter on days 1 and 2
The above trials use brain matter that has been centrifuged to generate a protein-free homogenate and a protein-rich homogenate based on molecular weight.
Rabbit 9 and 10: injected with boiled homogenized brain matter on days 1 and 2
The above trials use brain matter that have been boiled to destroy any bacterial contaminants in the homogenate.
In Rabbits 5 and 6, irradiation is used to destroy nucleic acids. What type of irradiation is likely to be used in this portion of the experiment?
Ionizing radiation, to generate free radicals.
Non-ionizing radiation, to prevent free radical generation
Ionizing radiation, to prevent free radical generation
All types of radiation have the potential to satisfactorily destroy nucleic acid
Non-ionizing radiation, to generate free radicals
Ionizing radiation, to generate free radicals.
Students should be familiar with ionizing radiation as the type of radiation that generates free radicals. Further, it should be understood that free radical formation causes hyper-reactivity among exisitng nucleic acid molecules and subsequent destruction.
Example Question #101 : Macromolecules
How many hydrogen bonds does it take to hold adenine and thymine together? Cytosine and guanine?
1; 3
1; 2
2; 4
2; 2
2; 3
2; 3
In order to hold a base pair together, the hydrogen bond(s) between the bases must be stable. This stability depends on many factors, such as the size and shape of the bases. The adenine-thymine base pair is most stable when held together by 2 hydrogen bonds, while the cytosine-guanine base pair is most stable when held together by 3 hydrogen bonds.
Certified Tutor
All MCAT Biology Resources





