All MCAT Biology Resources
Example Questions
Example Question #1 : Genetic Abnormalities And Mutation
Scientists use a process called Flourescent In-Situ Hybridization, or FISH, to study genetic disorders in humans. FISH is a technique that uses spectrographic analysis to determine the presence or absence, as well as the relative abundance, of genetic material in human cells.
To use FISH, scientists apply fluorescently-labeled bits of DNA of a known color, called probes, to samples of test DNA. These probes anneal to the sample DNA, and scientists can read the colors that result using laboratory equipment. One common use of FISH is to determine the presence of extra DNA in conditions of aneuploidy, a state in which a human cell has an abnormal number of chromosomes. Chromosomes are collections of DNA, the totality of which makes up a cell’s genome. Another typical use is in the study of cancer cells, where scientists use FISH labels to ascertain if genes have moved inappropriately in a cell’s genome.
Using red fluorescent tags, scientists label probe DNA for a gene known to be expressed more heavily in cancer cells than normal cells. They then label a probe for an immediately adjacent DNA sequence with a green fluorescent tag. Both probes are then added to three dishes, shown below. In dish 1 human bladder cells are incubated with the probes, in dish 2 human epithelial cells are incubated, and in dish 3 known non-cancerous cells are used. The relative luminescence observed in regions of interest in all dishes is shown below.
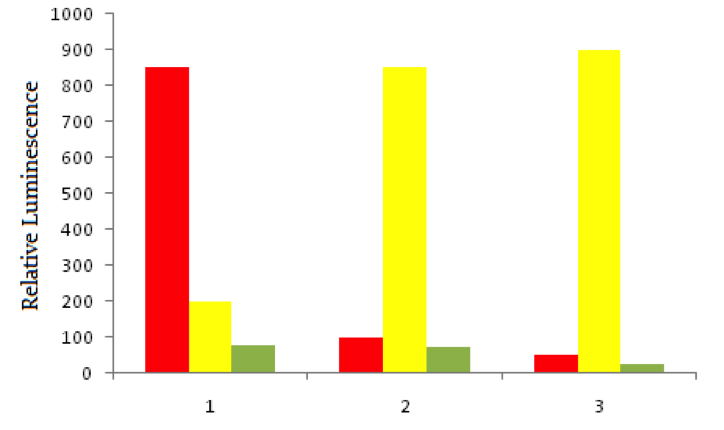
Which of the following genetic changes would not be detectable using FISH?
Single nucleotide polymorphism
Copy number variant
Robertsonian translocation
Balanced translocation
Unbalanced translocation
Single nucleotide polymorphism
FISH is useful for visualizing major changes in the genome, as described by the passage. Translocations and large copy number variants would likley be visible in color changes, while small single nucleotide polymorphisms (SNPs) would likely be less easily detected.
Example Question #1 : Genetic Abnormalities And Mutation
Which of the following represents a frameshift mutation to the given template strand?
5'-AGCCTTAGC-3'
5'-TTTAGCCTTAGC-3'
5'-AGCCTTAGG-3'
5'-AGCGCTTAGC-3'
5'-CTTAGC-3'
5'-TGCCTTAGC-3'
5'-AGCGCTTAGC-3'
A frameshift mutation results in a change of the codon reading frame and results from the addition or deletion of a set of nucleotides that is not a multiple of three. If a mutation occurs that is a multiple of three, the reading frame is unchanged and a simple addition or deletion has occurred.
Template: 5'-AGC-CTT-AGC-3'
Frameshift mutant: 5'-AGC-GCT-TAG-C-3'
Point mutant: 5'-TGC-CTT-AGC-3'
Point mutant: 5'-AGC-CTT-AGG-3'
Deletion: 5'-CTT-AGC-3'
Insertion: 5'-TTT-AGC-CTT-AGC-3'
Note that all except the frameshift mutation contain sets of three nucleotides to create triplets. The frameshift leaves a singular, un-grouped cytosine.
Example Question #1110 : Biology
Cryptosporidium is a genus of gastrointestinal parasite that infects the intestinal epithelium of mammals. Cryptosporidium is water-borne, and is an apicomplexan parasite. This phylum also includes Plasmodium, Babesia, and Toxoplasma.
Apicomplexans are unique due to their apicoplast, an apical organelle that helps penetrate mammalian epithelium. In the case of cryptosporidium, there is an interaction between the surface proteins of mammalian epithelial tissue and those of the apical portion of the cryptosporidium infective stage, or oocyst. A scientist is conducting an experiment to test the hypothesis that the oocyst secretes a peptide compound that neutralizes intestinal defense cells. These defense cells are resident in the intestinal epithelium, and defend the tissue by phagocytizing the oocysts.
She sets up the following experiment:
As the neutralizing compound was believed to be secreted by the oocyst, the scientist collected oocysts onto growth media. The oocysts were grown among intestinal epithelial cells, and then the media was collected. The media was then added to another plate where Toxoplasma gondii was growing with intestinal epithelial cells. A second plate of Toxoplasma gondii was grown with the same type of intestinal epithelium, but no oocyst-sourced media was added.
You are conducting a study of an isolated tribe in New Guinea, and you find that there is widespread resistance to cryptosporidium infection. Upon examination, you find that the resistance is caused by a change in one nucleotide pair in a gene on chromosome 13. What kind of genetic change does this likely reflect?
Genomic imprinting
Chromosomal change
Copy number variant
Fragile X
Single nucleotide polymorphism (SNP)
Single nucleotide polymorphism (SNP)
This would be an example of a single nucleotide polymorphism. A fairly common variant is some change at a single base pair in human DNA. It is possible that this base pair change results in a modified protein that functions just as well as the normal protein in most conditions.
Some of these changes may, simply by chance, be better at resisting disease. When exposed to stress, such as a disease epidemic, it is possible this variant of normal becomes the most widespread genotype. This is especially noticeable in an isolated population, like a tribe in New Guinea. Note that not all single nucleotide polymorphisms will have positive effects, and in some cases can cause disease instead of resistance.
Example Question #2 : Genetic Abnormalities And Mutation
Cellular division is an essential part of the cell cycle. When a cell divides it passes genetic information to daughter cells. The amount of genetic information passed on to daughter cells depends on whether the cell undergoes mitosis or meiosis. Mitosis is the most common form of cell division. All somatic cells undergo mitosis, whereas only germ cells undergo meiosis. Meiosis is very important because it produces gametes (sperm and eggs) that are required for sexual reproduction. Human germ cells have 46 chromosomes (2n = 46) and undergo meiosis to produce four haploid daughter cells (gametes).
An individual containing three sex chromosomes (XXY) is called a polysomic individual. What is the reason for polysomy?
Nondisjunction of chromosomes during meiosis
Fertilization of two sperm cells with two eggs
Presence of multiple polar bodies in a female after meiosis
Splitting of a single fertilized egg into two or more eggs
Nondisjunction of chromosomes during meiosis
The question states that the individual with three sex chromosomes has a condition called polysomy. Recall that a normal individual will only carry two copies of a chromosome.
This abnormality occurs when meiosis isn’t carried out properly. A daughter cell from meiosis can contain an extra chromosome if sister chromatids don’t separate properly during anaphase II (a process called nondisjunction). This extra chromosome can be carried over to the offspring, giving rise to a polysomic individual.
Fertilization of two sperm cells with two eggs gives rise to fraternal twins (non-identical twins) and splitting of a single fertilized egg into two or more eggs gives rise to monozygotic twins (identical twins); therefore, you can eliminate these two answer choices. After meiosis, females always possess multiple polar bodies. During meiosis in females, most of the cellular content is transferred to a single daughter cell: the egg. The remaining daughter cells contain the remnants and are called polar bodies. These polar bodies don’t participate in development and fertilization.
Example Question #62 : Cell Biology, Molecular Biology, And Genetics
Chromosomal aberrations, such as trisomy or monosomy, are often the result of nondisjunction during cell division. Nondisjunction is characterized by a malfunction during which stage of division?
Anaphase
Telophase
Metaphase
Interphase
Anaphase
This question is asking about the phase of Meiosis, during which disjunction (separation) of chromosomes normally occurs. While Metaphase can sometimes be a seductive answer, note that disjunction officially occurs during Anaphase.
Example Question #2 : Genetic Abnormalities And Mutation
An mRNA sequence is supposed to read UAUGGA, but a mutation replaces the second uracil base with guanine. What is the most specific term for this type of mutation?
Missense
Nonsense
Insertion
Frame-shift
Deletion
Nonsense
This mutation replaced UAU (a coding codon) with UAG (a stop codon). The most descriptive term for this kind of replacement is a nonsense mutation.
Note that an easy way to remember the stop codons is UGA ("u" get away), UAA ("u" are away), and UAG ("u" are gone).
Example Question #1 : Genetic Abnormalities And Mutation
A mutation within a gene results in the premature addition of a stop codon during translation. This describes which type of mutation?
Nonsense
Missense
Degradation
Frameshift
Nonsense
A nonsense mutation results in premature addition of a stop codon. A missense mutation is a point mutation that results in a different codon, which ultimately codes for a different amino acid in the polypetide sequence. A frameshift mutation results from and insertion or deletion of a nucleotide, resulting in a change in the reading frame. Frameshift mutations often indirectly result in nonsense mutations, but are not the best answer choice given.
Example Question #1112 : Biology
Imagine there is a mutation in a gene where a nucleotide was replaced by another nucleotide. The mutation did not affect the primary structure of the protein for which the gene coded.
What type of mutation is this?
Frameshift mutation
Insertion mutation
Point mutation
Nonsense mutation
Point mutation
Whenever a base pair is replaced by another base pair in the DNA double helix, it is referred to as a point mutation. If the amino acid is not changed by the mutation, we can refer to the mutation as a silent mutation, as the primary protein structure is not affected.
Point mutation: CATGA becomes CAGGA
Insertion mutations occur when additional bases are inserted into the DNA sequence.
Insertion mutation: CATGA becomes CATACTGA
Frameshift mutations can result from insertions or deletions when the number of nucleotides added/removed is not a multiple of three. Since codons are grouped by threes, any change that is not a multiple of three will alter the grouping of every codon downstream of the mutation, severly altering the primary protein structure.
Frameshift mutation: CATGA becomes CAATGA
A nonsense mutation results in a premature stop codon, and early translation termination. This can arise from a frameshift mutation, point mutation, insertion, or deletion.
Nonsense mutation: CATGA becomesCATTTAGA (When transcribed, this sequence becomes the mRNA UCUAAAUG, where UAA is a stop codon).
Example Question #1111 : Biology
Consider the pedigree. Is the trait dominant or recessive?
Recessive
Dominant
The mode of inheritance cannot be determined
Codominant or incompletely dominant
Recessive
The trait is recessive because affected individuals do not occur in every generation. Additionally, dominant traits do not result in "carriers". Individuals are either affected or unaffected.
Example Question #72 : Genetics
The concept of genomic imprinting is important in human genetics. In genomic imprinting, a certain region of DNA is only expressed by one of the two chromosomes that make up a typical homologous pair. In healthy individuals, genomic imprinting results in the silencing of genes in a certain section of the maternal chromosome 15. The DNA in this part of the chromosome is "turned off" by the addition of methyl groups to the DNA molecule. Healthy people will thus only have expression of this section of chromosome 15 from paternally-derived DNA.
The two classic human diseases that illustrate defects in genomic imprinting are Prader-Willi and Angelman Syndromes. In Prader-Willi Syndrome, the section of paternal chromosome 15 that is usually expressed is disrupted, such as by a chromosomal deletion. In Angelman Syndrome, maternal genes in this section are deleted, while paternal genes are silenced. Prader-Willi Syndrome is thus closely linked to paternal inheritance, while Angelman Syndrome is linked to maternal inheritance.
Figure 1 shows the chromosome 15 homologous pair for a child with Prader-Willi Syndrome. The parental chromosomes are also shown. The genes on the mother’s chromosomes are silenced normally, as represented by the black boxes. At once, there is also a chromosomal deletion on one of the paternal chromosomes. The result is that the child does not have any genes expressed that are normally found on that region of this chromosome.
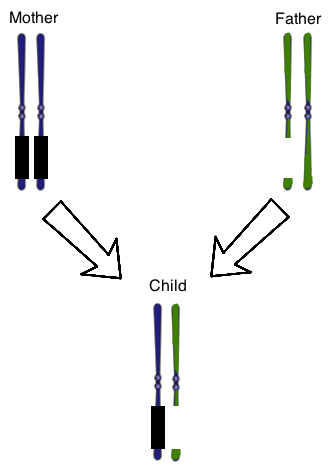
In addition to the chromosomal deletion on chromosome 15 in the passage, the father is found to have another gene with a mutation, which adds a stop codon prematurely in the base pair sequence. This mutation is best described as a __________.
frameshift mutation
non-conservative missense mutation
conservative missense mutation
silent mutation
nonsense mutation
nonsense mutation
The best answer is a nonsense mutation, which is defined as a point mutation that gives rise to an early stop codon, thus truncating any protein products prematurely. These are typically devastating mutations for protein function.
Certified Tutor
All MCAT Biology Resources